How to import KEGG pathway xml(kgml) to Cytoscape¶
First we show how to import KEGG pathway xml(kgml) to Cytoscape.
Importing kgml to Cytoscape with REST endpoint¶
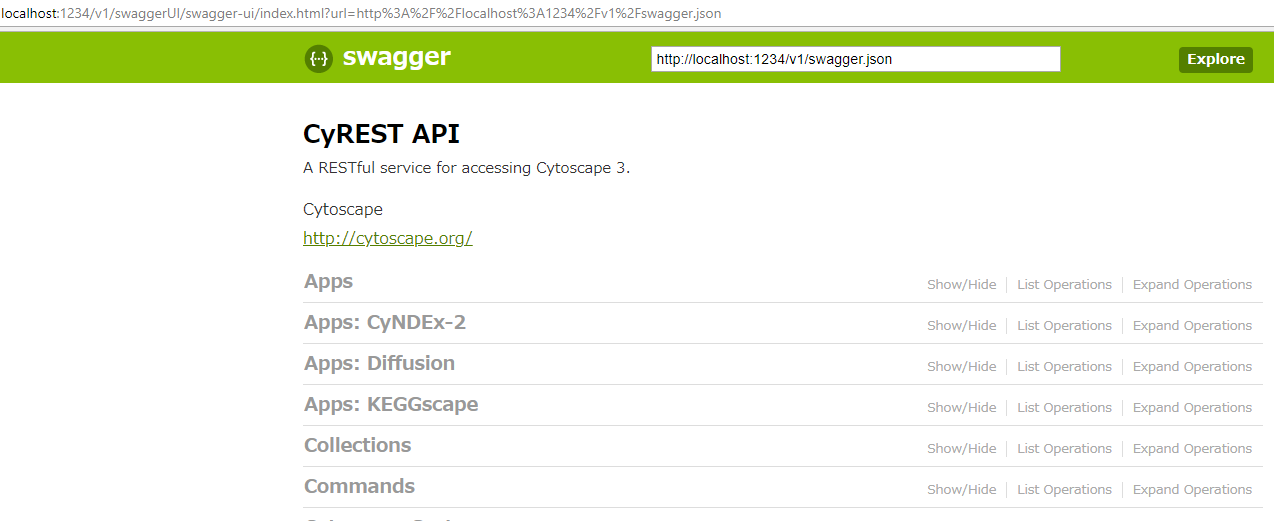
KEGGscape exposes a REST endpoint to directly import a KEGG pathway entry and it is documented in the main Swagger page generated by CyREST (available under: Help -> Automation -> CyREST API).
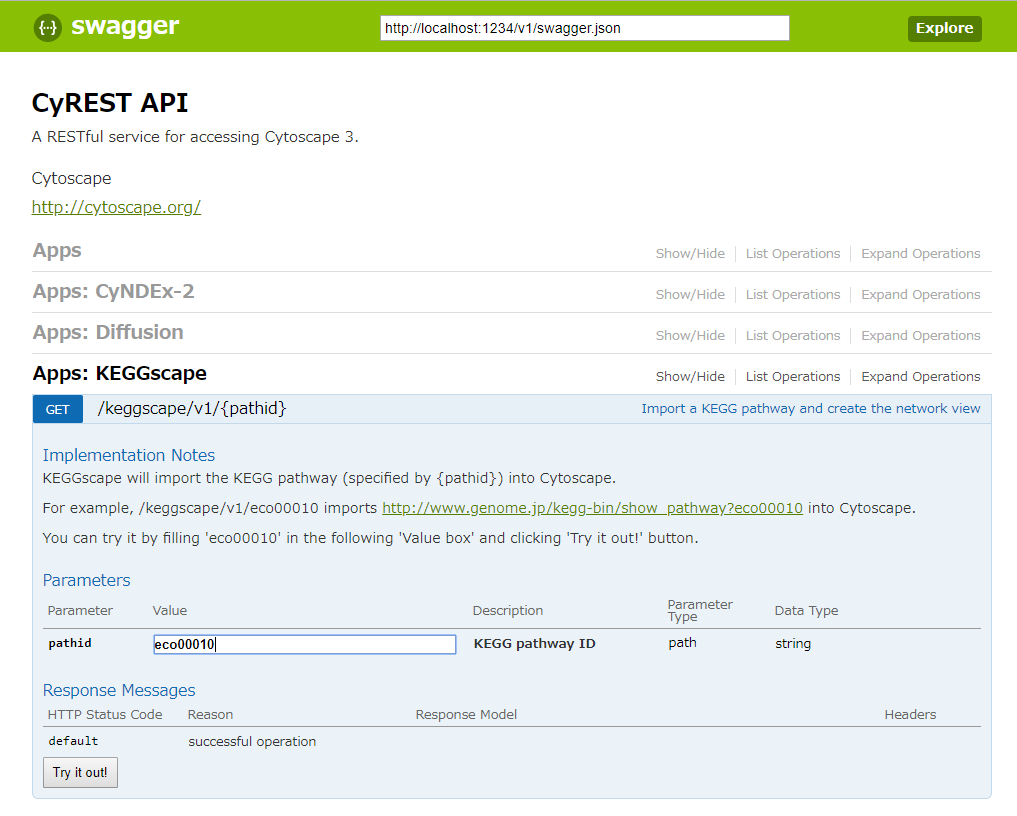
You can import kgml to Cytoscape with filling the KEGG pathway ID and clicking the “Try it out!” button.
Importing kgml to Cytoscape by manually downloading kgml¶
Downloading KEGG pathway kgml¶
You can download KEGG pathway kgml without opening web browser (if you know the KEGG pathway entryID you want to import).
wget http://rest.kegg.jp/get/eco00020/kgml -O eco00020.xml
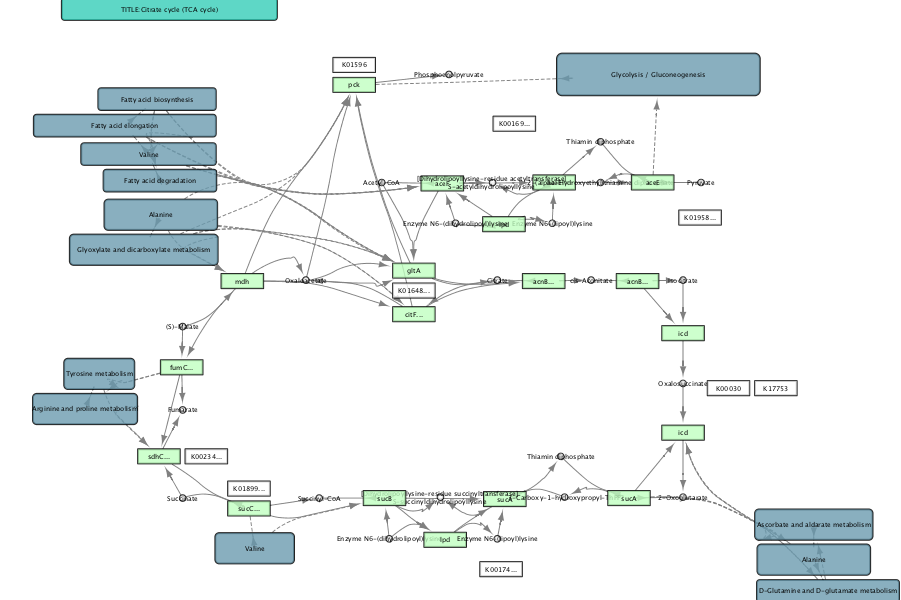
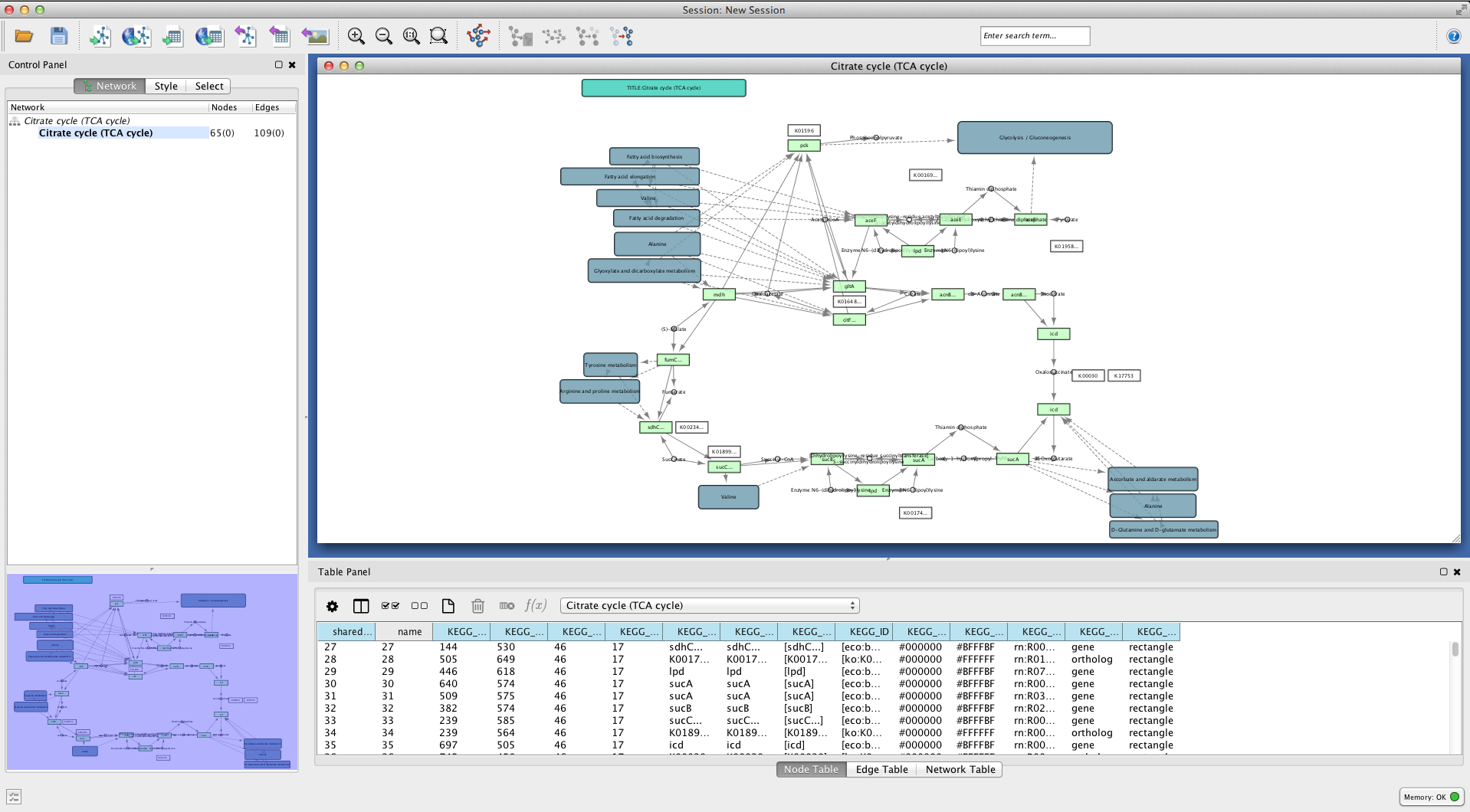
eco00020.xml is TCA cycle of Escherichia coli K-12 MG1655.
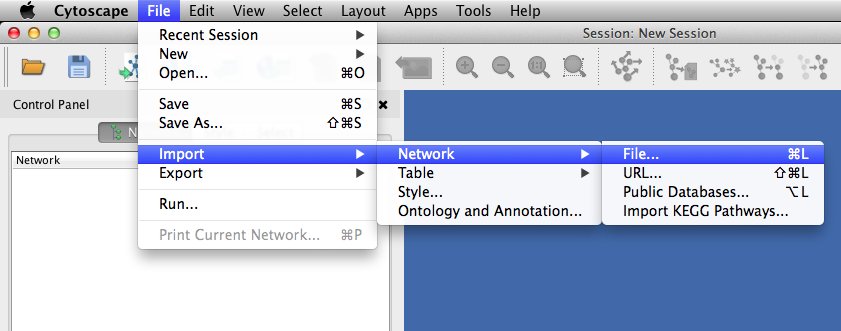
Importing kgml to Cytoscape by GUI¶
You can import kgml to Cytoscape from menu bar
File -> Import -> Network -> File
and open eco00020.xml.
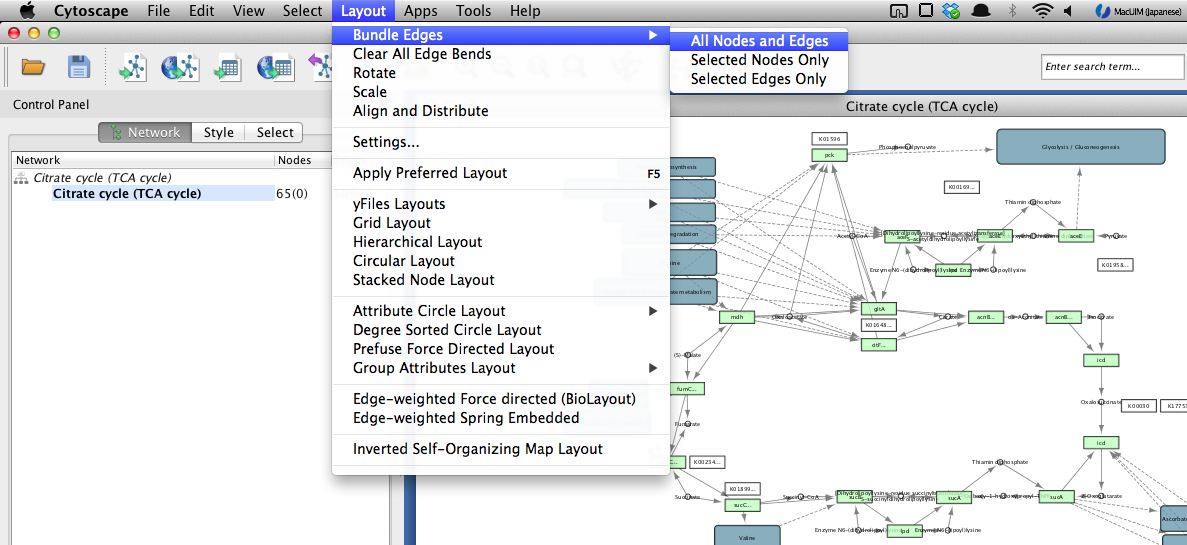
How to bundle edges¶
KEGGscape creates two edges for a reversible reaction, if you want to bundle these reversible reactions like KEGG, please select “Bundle edges” from “Layout” menu.